Hyphochytrium catenoides Genome Sequencing
"Eukaryotic microbes have three primary mechanisms for obtaining nutrients and energy: phagotrophy, photosynthesis and osmotrophy. Traits associated with the latter two functions arose independently multiple times in the eukaryotes. The Fungi successfully coupled osmotrophy with filamentous growth, and similar traits are also manifested in the Pseudofungi (oomycetes and hyphochytriomycetes). Both the Fungi and the Pseudofungi encompass a diversity of plant and animal parasites. Genome-sequencing efforts have focused on host-associated microbes (mutualistic symbionts or parasites), providing limited comparisons with free-living relatives. Here we report the first draft genome sequence of a hyphochytriomycete ‘pseudofungus’; Hyphochytrium catenoides. Using phylogenomic approaches, we identify genes of recent viral ancestry, with related viral derived genes also present on the genomes of oomycetes, suggesting a complex history of viral coevolution and integration across the Pseudofungi. H. catenoides has a complex life cycle involving diverse filamentous structures and a flagellated zoospore with a single anterior tinselate flagellum. We use genome comparisons, drug sensitivity analysis and high-throughput culture arrays to investigate the ancestry of oomycete/pseudofungal characteristics, demonstrating that many of the genetic features associated with parasitic traits evolved specifically within the oomycete radiation. Comparative genomics also identified differences in the repertoire of genes associated with filamentous growth between the Fungi and the Pseudofungi, including differences in vesicle trafficking systems, cell-wall synthesis pathways and motor protein repertoire, demonstrating that unique cellular systems underpinned the convergent evolution of filamentous osmotrophic growth in these two eukaryotic groups."
Comparative genomic analysis of the ‘pseudofungus’ Hyphochytrium catenoides. (2018). Leonard et. al. PNAS. https://dx.doi.org/10.1098%2Frsob.170184
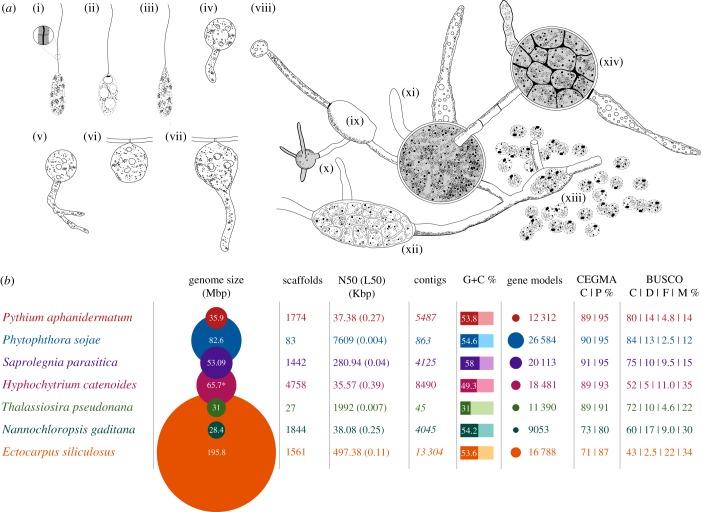
Figure - Developmental characteristics of H. catenoides and genome statistics of representative stramenopiles. Sketches of a subset of different stages of H. catenoides life cycle, adapted and redrawn from [6,14] showing: (i–iii) different views of zoospores (including magnification of tinselate flagellum i), (iv) germination stage of large spore, (v) primary enlargement or primary sporangium, (vi,vii) thallus development on substrate, (viii) unusual extensive branched thallus, which consists of separated sporangia at different stages of maturity (e.g. xii,xiv), connected by long, tubular, septate, hyaline and empty hyphae (x,xi), sometimes with enlargements without sporangia (e.g. ix). Zoospores may fail to swim coming to rest near exit tube (xiii). (b) Table of genome statistics for a range of different stramenopiles. Asterisk indicates k-mer estimation of genome size (column 2). All numbers are from the respective genome datasets (see electronic supplementary material, table S12). Numbers in italics (contigs, column 5) are inferred from the scaffolded data. CEGMA: C, complete; P, partial recovered gene models. BUSCO: C, complete; D, duplicated; F, fragmented; M, missing gene models.
Links
- Genome Resources on GitHub
- Genome Assembly at NCBI
- BLAST Server - Coming Soon!